I have the privilege to work in the lab of Steven Kushner and Femke de Vrij, within the Department of Psychiatry at the Erasmus Medical Center in Rotterdam. Our lab focuses on neuropsychiatric disorders of the brain, like Schizophrenia and Angelman syndrome. Because the brain cannot simply or ethically be sampled in a live human, we study models that are as similar as possible to circumvent this issue, like animals and cell cultures. In the past couple of decades researchers discovered a way to reprogram somatic cells into stem cells, hereby creating induced pluripotent stem cells (iPSC). Because we can now take somatic cells from patients specifically, we can study these cells while they retain the genetic background, instead of having to rely on cell lines or animals. This has been revolutionary to the field and has allowed more direct access to study living human brain cells in vitro, with the dream of elucidating neurobiological mechanisms of brain diseases.
My projects are aimed at bioinformatic analysis aligned with multiple projects in the lab. For instance, the analysis of single-cell RNA sequencing data for different iPSC-derived neural differentiation models together with Bas Lendemeijer and Mark van der Kroeg in the lab. For these models, such as a novel iPSC-derived astrocyte protocol and adherent cortical organoids, we are interested to know the cell-type specific expression patterns and how they compare developmentally to living human brain tissue. To get to this understanding, we use single-cell RNA sequencing for looking at the transcriptomic expression profiles and analyzing these data. For this, I am developing a pipeline in the R statistical programming language with the Seurat framework that enables data processing and analyses, visualization and statistics in order to generate results which we can then examine together.
To elaborate on my project specifically, Bas Lendemeijer has worked on protocols to differentiate astrocytes, neurons by NGN2 overexpression. These monocultures can then be co-cultured, from all of these cultures we can take samples and ready them for RNA sequencing. Here we need to think about what type of RNA to sequence, RNA purification and enrichment, generating complementary DNA libraries and more. After sequencing we gain sequences which we align to a reference genome for mapping to genes and counting the number of matches. This raw data is then preprocessed for quality control, cells and genes are filtered for optimizing downstream processing. Normalization is performed to reduce batch effects and to allow comparisons between samples. Often a principal component analysis is used as a technique for dimensionality reduction in order to retain most of the variance in the data within fewer features. From here cells are clustered to see how they separate based on their genetic profiles, these clusters are then annotated for cell type identification. With these annotated profiles, and while using all the count data again, we then perform differential expression analyses to statistically identify genes that contribute most of the variation to certain groups. These significant genes can then be used for functional analyses like lineage tracing, cell-cell interaction networks, gene ontology/set enrichment in order to hopefully elucidate biological mechanisms.
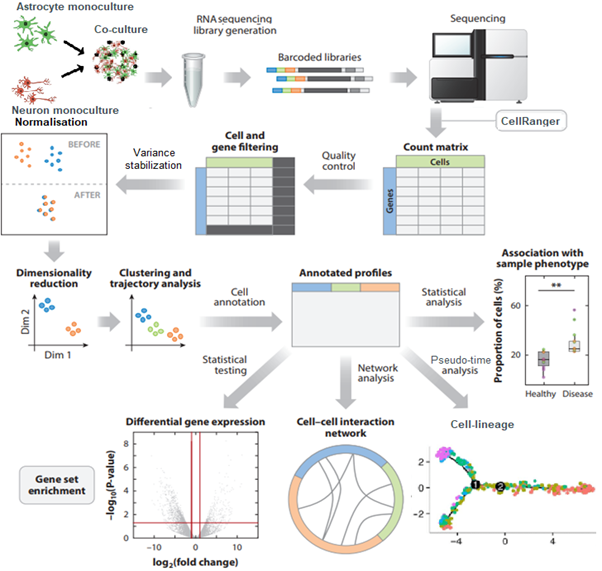
Common single-cell RNA sequencing analysis pipeline annotated with popular approaches and tools. Figure adapted from a publication by Brian Hie et al.: https://doi.org/10.1146/annurev-biodatasci-012220-100601
Being a bioinformatician in a lab with predominantly biologists, I like to focus on bridging this biological and technical aspects. Which requires communication to link expertise and jargon and from different niches to come to an understanding together. Within the Netherlands Organ on Chip Initiative (NOCI) this has been of great importance and delight as well. This network consisting of people with multidisciplinary backgrounds that share knowledge and work together has been amazing to be part of. Facilitating collaboration through creativity and possibilities with multidisciplinary projects that together are larger than the sum of their parts, strengthened by the power of combining disciplines.
What we thought was still science fiction not so long ago has since become reality, the combined technical and biological revolutions of the past decades have catapulted us into a time of rapid advancement and allowing us to dream big. I am extremely excited for the future with all the new discoveries to be made, speeding towards an era of personalized medicine and increased well-being for everyone.

Maurits Unkel
PhD candidate at EMC
