One of the areas that NOCI is investigating is the development of brain-on-a-chip models for epilepsy. I was recently invited to write a Dutch piece in the journal “Epilepsie, periodiek voor professionals”, where we explain how we can replicate and analyze complex epileptic electrophysiological recordings using brain on-a-chip systems and automated analytical softwares (https://epilepsiejournal.nl/article/view/11399). This was an interesting topic and I thought I could share it here as well, albeit in English.
Introduction
The analysis of large data sets has become an interesting addition to the repertoire of scientific research. Nowadays, data acquisition is faster and storage is easier, resulting in a fast amount of data that calls for efficient analysis.1 For instance, in epilepsy studies, neurophysiological recordings easily result in gigabytes of data due to a high temporal and spatial resolution. In basic research, protocols to systematically extract a disease signature are currently lacking. Also, custom-made analysis protocols may introduce variability in interpretation if processing, analyzing and post-processing of data are incompletely transmitted. This is particularly the case in academic studies where neuroscientists with limited coding expertise need to evaluate the electrophysiological output of their neuronal cultures. Therefore, tools to help analyze complex data sets efficiently (i.e: 20,000 data points per second per electrode, ranging from 12 to 1000 electrodes depending on the MEA layout and type, that needs to be analyzed in correlation with one another for every data point), in a user-friendly and standardized manner are needed to enable the field to move forward.
Experimental model with human biopsy and analysis
Traditionally, most of the experimental models in epilepsy are based on animals. However, recent focus has shifted to explore human induced pluripotent stem cells (hiPSC)2 as recent societal debate pushed the scientific community to use less animals in their research.3 hiPSCs have two advantages: Firstly, the models are based on human, not rodent, cells. This may contribute to overcoming the fact that over 90% of human drug trials are unsuccessful if they are primarily based on animal experimentation4. Thus, findings from animal models may not translate to patients despite both species sharing many similarities. The poor translational value may furthermore come from the fact that animal models typically represent a certain aspect of human pathophysiology and that readouts in animal research are different than in clinical research (poor external validity).
Secondly, a notable advantage of hiPSCs is that they can be derived from patients with a specific gene mutation so contain the exact genetic background of the patient. When combined with organ on-a-chip (OoC) technology, the model can be fine-tuned to mimic any type of organ, including brain-like structures. These advancements are clearly paving the way for personal medicine types of treatments.
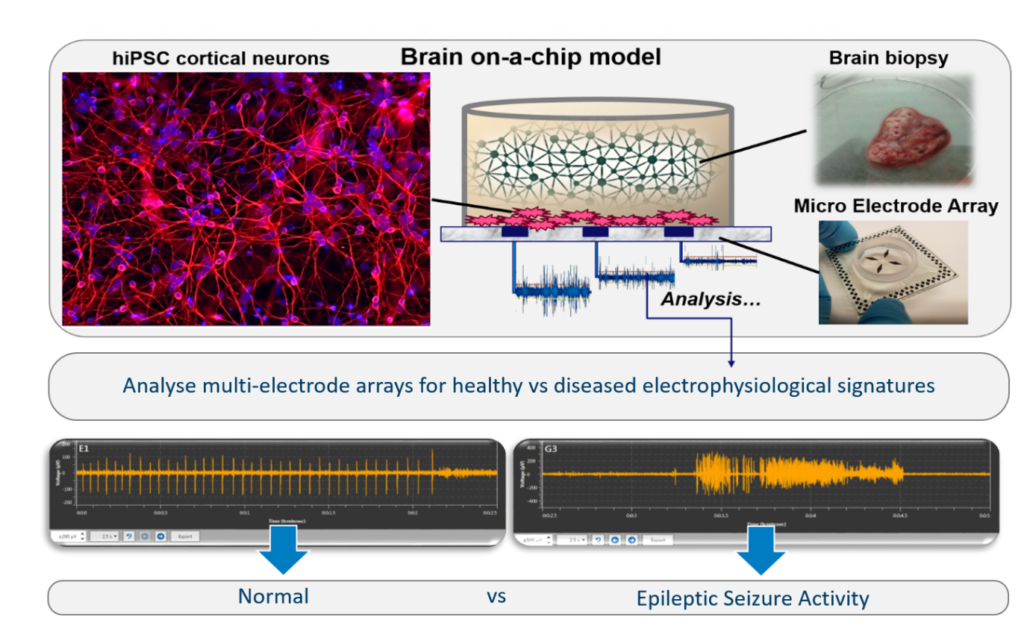
Figure 1: Brain on-a-chip model for in vitro epileptic electrophysiological recordings.
In a collaboration between Leiden University Medical Center (LUMC), Maastricht University Medical Center+ (MUMC+) and Technical University of Eindhoven (TU/e), we are developing our models for in vitro epileptic studies using hiPSC cells combined with a human neocortical biopsy (Figure 1), collected during epilepsy surgery. The model is coupled to a multi-electrode array (MEA) to record electrophysiology and monitor epileptic activity of the neuronal cells. The MEA consists of an array of microscopic electrodes on which the electrical behavior of neurons can be recorded and is already a popular tool for disease modeling and drug screening.5 However, the amount of data that is collected during MEA recordings is large (no less than ~1 GB per minute of recording of 120 electrodes) and complex to understand without using advanced algorithms and coding expertise, which are often not accessible to neurobiologists. Because each data set is unique, there is currently no standardized method to analyze these rich MEA data sets, so most neurobiologists use unpublished in-house developed scripts to analyze their data. To fill a crucial void, we decided to create an automated software called “MEAtoolbox” that will allow users to analyze their MEA data without the need for any coding expertise and provide them with multiple tools to analyze the data set in an effort to standardize the MEA analysis pipeline. Interestingly, in another collaboration within NOCI involving TU/Delft (Dr Massimo Mastrangeli), we are developing 3D MEAs to record electrophysiology from organoids and other 3D cellular constructs, and we have made our MEAtoolbox compatible with such 3D chip MEA devices already.
Discussion
The MEAtoolbox (figure 2A) is built on three main foundations of tools that will allow the user to automatically detect and analyze spikes, bursts and network bursts. Furthermore, we also included tools that allow the detection and analysis of functional connections as well as a function for spike sorting. All of the tools are packaged in a single, user-friendly, graphical user interface (GUI) that allows users to analyze MEA data instantly (figure 2B). After all the files are analyzed, the toolbox will open a GUI and allow the user to explore the analyzed data and fine-tune the data if the user disagrees (figure 2C). Once the post-processing is finished, the user can export the results with the click of a button. The results is an Excel file with 20 endpoints (e.g. firing rate, burst number and duration, inter-spike intervals, inter-burst intervals, network activity) that provide information about spike train metrics, burst metrics and network burst metrics (figure 2D). MEAtoolbox was used first to characterize our epileptic model that consists of a MEA with hiPSCs combined with a human neocortical biopsy obtained after surgery. Clear epileptic form of bursting activity was observed during the live measurements for 6 consecutive days, which is much longer that the few hours normally used for such experiment. Such characterization of the electrophysiological signature was instead very challenging to extract correctly and -in a way- less time-efficient manner using existing software. Instead, we found that MEAtoolbox could efficiently extract all required information by pressing a few buttons in the GUI and due to its automation capabilities, MEAtoolbox could analyze the whole data set in one go, whilst automatically providing information about spikes, bursts and network bursts. Moreover, using MEAToolbox, we could identify that the electrophysiological signatures obtained in our epileptic model matched with a seizurogenic signature5. This signature can be characterized by an increase in fire rate, burst rate, burst duration, coefficient of variation of inter-spike interval (ISI) and network burst durations rate and a decrease in isolated spikes and inter-spike intervals.
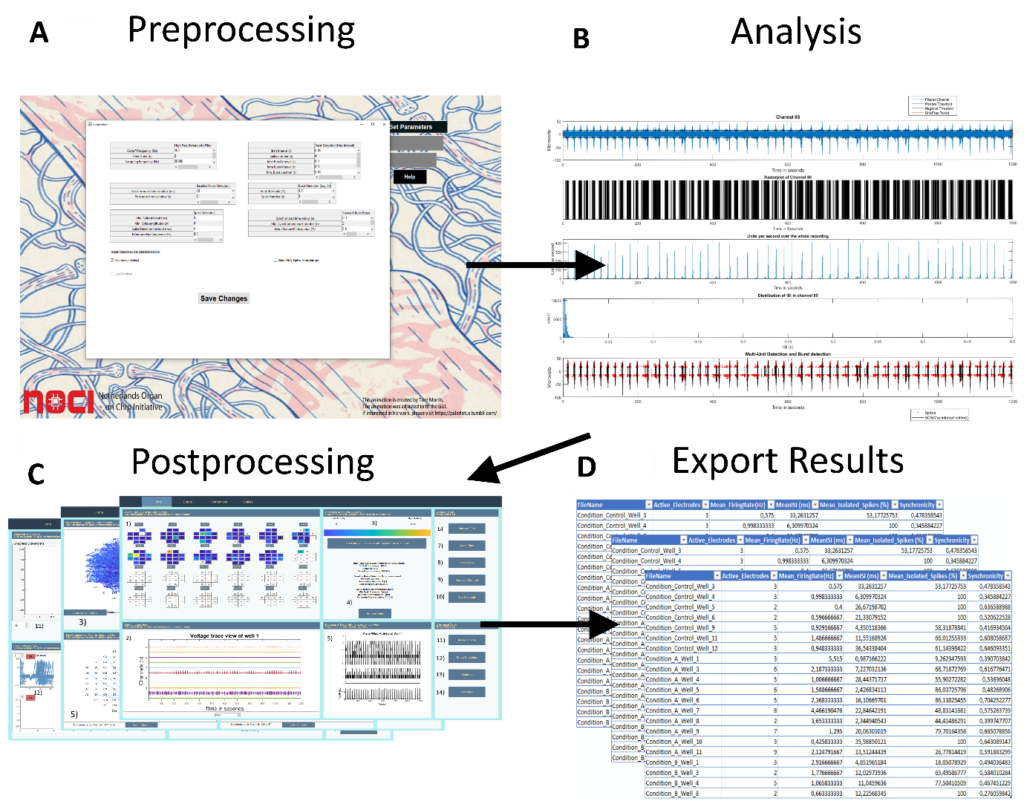
Figure 2: The analysis pipeline of the MEAToolbox to analyze MEA data.
Conclusion
We have created an effective toolbox that will help neurobiologists with the analysis of MEA data in a more standardized manner without the need of coding expertise. We do realize that the toolbox is not perfect or suited for all types of MEA experiments – since all MEA data is unique -, making full standardization particularly challenging. However, MEAtoolbox is free, open source and available on our github page (https://github.com/mhyhu/Toolbox) so experts in signal processing and electrophysiological analysis can access the source code and add to it to further improve standardization and refine some of the algorithms to suit more needs. In conclusion, we show that the MEAtoolbox can be a valuable tool to interpret complex electrophysiological signals from, for instance, an epileptic human brain biopsy to identify pathological signatures in vitro.
References
- Amado, A., Cortez, P., Rita, P. & Moro, S. Research trends on Big Data in Marketing: A text mining and topic modeling based literature analysis. Res. Manag. Bus. Econ.24, 1–7 (2018).
- Takahashi, K. & Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell126, 663–676 (2006).
- Hubrecht RCand Carter E. The 3Rs and Humane Experimental Technique: Implementing Change. Animals (Basel). 2019 Oct; 9(10): 754. doi: 3390/ani9100754.
- AYSHA AKHTAR. The Flaws and Human Harms of Animal Experimentation. Camb Q Healthc Ethics. 2015 Oct; 24(4): 407–419. doi: 10.1017/S0963180115000079.
- Bradley, J. A., Luithardt, H. H., Metea, M. R. & Strock, C. J. In vitro screening for seizure liability using microelectrode array technology. Sci.163, 240–253 (2018).

Jean-Philippe Frimat
NOCI Postdoc at LUMC
